Glasner Lab
Immunology and Immunotherapy
We are studying immune-stromal communication from basic science to translational applications
The immune system is primarily known for its role in defending the body against invading pathogens. However, immune cells also play a crucial role in maintaining and regulating tissue homeostasis. To effectively perform this dual function, the immune system must carefully balance the activation and suppression of various cells, pathways, and mediators. These extensive interactions between immune cells and stromal cells in different tissues are understudied, and they were especially overlooked so far in the context of pathology. It is our hypothesis, that immune cells perform diverse supportive functions in the tissues regardless of their classic immune functions, and by studying these unexplored pathways we aim to design new targets for immunotherapy.
Single-cell sequencing
We use autochthonous, genetic, and orthotropic models to explore tumor development and other types of model pathologies, such as inflammation-induced lung injury, as well as samples from human patients to study the interactions between various cells and tissues. Using these models, and our laboratory-specialized tissue slice culture system, we compare depletion or over-expression conditions with untreated controls using methods such as single-cell sequencing to analyze the gene expression profiles of individual cells within the tissue or tumor microenvironment (TME). This type of analysis generates a comprehensive understanding of the complex network of interactions within the tissue or TME. Detailed transcriptomic profiling of the entire spectrum of cellular components in a tissue offers insights into the roles of specific immune cells in the development and progression of tumors or other pathologies and can help identify pathways that may be targeted for therapeutic intervention.
Spatial Transcriptomics
Spatial transcriptomics is a technology that enables us to analyze the spatial organization of gene expression features in tissues at a high resolution by sequencing of barcoded RNA molecules on a slide. The resulting data can be used to create detailed maps of gene expression within the tissue, providing insights into the function of different cell types and how they interact within the tissue. We observed that regulatory T cells (Tregs) tend to concentrate around the edges of tumor nodules in the lung and exist in closer proximity to stromal cells in these areas compared to tumor-free areas of the same lung. with ST we will examine the gene expression profiles of the various cell populations in the TME before and after perturbation, to determine how these profiles are influenced by cellular localization. This analysis will elucidate the mechanisms behind the effects of immune cells on tissue function and identify primary and secondary order effects, and which programs are cell intrinsic or influenced by the surrounding tissue environment. This approach gives us a unique and unparalleled view into these interactions and has the potential to revolutionize our understanding of the immune system and its role in different contexts or tissue-states.
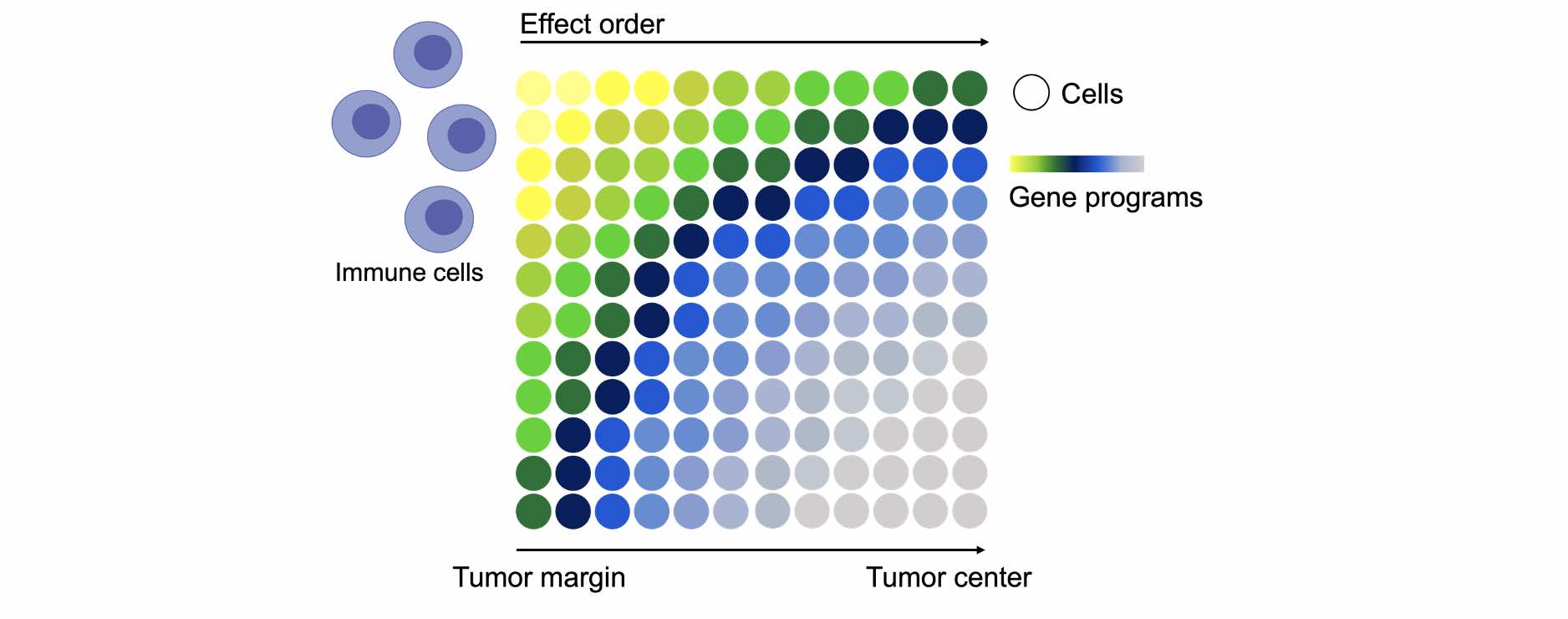
A schematic illustration of the types of questions that may be tackled by ST analysis. Cells are portrayed as circles, programs are presented as a colorful gradient. We will study the different programs employed by different types of cells in different parts of the tissue.
High throughput imaging
we also utilize high-throughput imaging to complement our ST experiments on the protein level. This allows us to view cell interactions in situ and gain a more comprehensive understanding of these complex processes. High throughput imaging assays allow us to detect multiple cell markers simultaneously, providing single-cell resolution and precise characterization of stromal cells and immunocytes on the protein level. By combining ST experiments with high throughput imaging, we are able to gain a more complete picture of immune stromal cell interactions and how they contribute to the overall function of the immune system in normal and pathogenic conditions.
The knowledge gained from our single-cell sequencing, ST, and imaging data can be used to dissect the complex network of interactions in tissues or the TME, and to potentially identify pathways that may be elicited as a secondary response to the original perturbation or treatment. By analyzing these data, we can gain a more comprehensive understanding of the complex network of interactions that occur within tissues and in the TME, and potentially identify pathways that may be targeted to improve current treatments or design new combination therapies.
Epigenetic profiling
Methods such as ATAC sequencing (a technique that allows us to map the accessibility of the genome, revealing which regions of DNA are open and available for transcription), ChIP-sequencing, and Cut&Run (methods for mapping protein-DNA interactions that involve cutting DNA with a specific enzyme and then analyzing the resulting fragments), can be used to study chromatin states and modifications, and the binding of transcription factors to enhancer elements or other sites in an organism genome. Epigenetic events can regulate gene expression and play a crucial role in the development and function of cells and tissues. By examining epigenetic marks, we can gain insight into the underlying mechanisms that control gene expression and how it is altered in different contexts. We use epigenetic profiling to complement single-cell sequencing, ST, and imaging for the purpose of building a holistic, comprehensive atlas of all levels of effects and interactions on tissues or the TME. By combining these techniques, we can gain a more complete understanding of the complex interplay between different genetic and epigenetic factors that shape the behavior of cells and tissues.
Parallel mouse and human tissue slice culture system
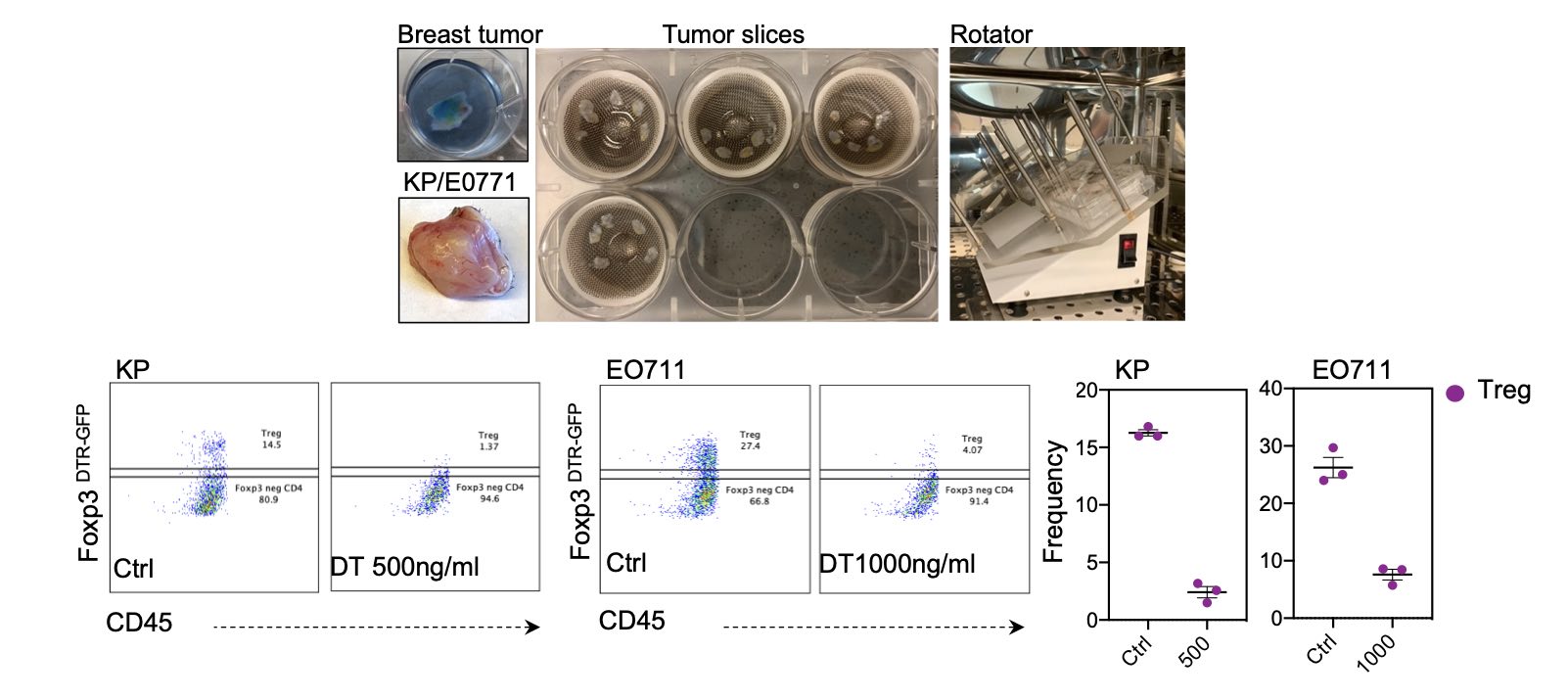
One way we study the effects of targeting specific pathways on tissue cell states and immune responses is to use an in-vitro tissue slice culture system, where mouse and human tissues are seeded and experimented on in culture. The sliced tissues retain many of the features and functions of the intact tissue for several days. This approach allows us to study the effects of perturbation on the immune system and other cells in the tissue in a controlled, experimental setting and on a small scale, allowing us to experiment with various treatments rapidly and simultaneously.
To use this approach, we obtain slices of mouse or human tissues and seed them in our lab-specialized slice culture system. In this system, tissues are sliced to a well-calibrated thickness and are placed on an angular rotator that allows for suitable gas exchange and media nutrient absorption. This allows us to study the effects of targeting specific pathways in a controlled, experimental setting, as a prerequisite before going into in-vivo studies using mouse models.
Importantly, seeding mouse and human tissues in parallel will allow us to focus only on conserved, clinically relevant pathways.
Generation and production of fusion proteins
The experiments that have been conducted thus far will provide us with insights into the various processes occurring in pathogenic tissue at different locations and levels of regulation. These processes may include pro- and anti-tumorigenic and primary and compensatory programs, which we will study using advanced statistical analysis. After combining these partially overlapping lists, we will identify a number of conserved pathways that could be potential targets for combination therapy. At this stage, we expect to have a carefully curated list of potential intervention targets, which we will then rank based on their effectiveness across programs, the level of modulation following an intervention, and their conservation between mice and humans. Some of these targets may be easily targeted with available commercial options, while others may be unknown, or difficult to target due to a lack of blocking/depleting antibodies or other molecular complexities and may require the creation of targeting constructs.
To this end, we will engineer fusion proteins (proteins that are made by cloning together two or more parts of separate proteins) using plasmids that carry a mutated fragment of the FC part of an antibody that is then fused to the receptor or binding region of the candidate protein. These fusion proteins will then be transfected into cells, expanded harvested, and purified to be used for blocking interactions in-vivo. These blocking interactions may occur by disrupting the function of the proteins they are engineered to bind. For example, if the mutated FC fragment is fused to the complement binding sequence of a signaling protein, the resulting fusion protein can bind to the receptor and block it from interacting with its natural ligand.
HSC transduction and transplantation
Following tissue slice culture experimentation ex-vivo, we will have pinpointed several robust, conserved pathways to further explore. To that end, we will generate mouse models for testing the effects of targeting these pathways for immunotherapy in-vivo.
As we want to screen many different pathways rapidly and efficiently, we will use bone marrow transplantation of cultured murine hematopoietic stem cells (HSCs) that will be modified ex-vivo. This procedure involves isolating HSCs from the bone marrow of mice, culturing the cells to allow for the introduction of genetic modifications, and then transplanting the modified HSCs into irradiated hosts. Once the modified HSCs have been transplanted and engrafted into the mice, they will give rise to hematopoietic cells that carry the genetic modifications. By using this approach, we can test the effects of targeting the pathways revealed in our previous high throughput screens in a controlled, experimental setting. This can provide valuable insights into the mechanisms underlying immune responses and the potential therapeutic benefits of targeting specific pathways for the treatment of various immune-related conditions.
In-vivo models
We have shown that the integration of all assays and methods employed thus far can provide insight into various unexplored aspects of TME immunity and suggest pathways for rational targeted combination therapy. We observed significant alterations in various gene programs following the short-term depletion of Tregs in a model of non-small cell lung carcinoma (NSCLC) in mice. One of the most upregulated pathways in several endothelial and mesenchymal subsets we observed following Treg cell depletion was the VEGF signaling pathway, which we identified as a promising targeting candidate for combination therapy. In a subcutaneous implementation model, we found that the combination of VEGF blockade and Treg cell depletion was more effective at restricting tumor growth than either approach alone or PD1 checkpoint blockade. These very promising results only scratch the surface of novel combination therapies that can be designed by studying and integrating the knowledge gained from our various lab projects.
Once we will have generated the relevant modified mice, we will utilize our autochthonous and orthotopic models (as well as other challenges) to ultimately test our candidate interventions in a specific tissue type as a last pre-clinical step. By using these in-vivo mouse models, we can simulate the conditions that occur in humans and study the effects of our candidate interventions on tumor development or other pathologies in a well-controlled and effective setup.
We are recruiting
Looking for driven, curious, hardworking and ambitious people who are excited about research